Virus
A virus is a small infectious agent thatreplicates only inside the living cells of otherorganisms. Viruses can infect all types of life forms, from animals and plants tomicroorganisms, including bacteria andarchaea.[1]
Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants, and the discovery of thetobacco mosaic virus by Martinus Beijerinckin 1898,[2] about 5,000 virus species have been described in detail,[3] although there are millions of types.[4] Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity.[5][6]The study of viruses is known as virology, a sub-speciality of microbiology.
While not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles. These viral particles, also known as virions, consist of two or three parts: (i) the genetic materialmade from either DNA or RNA, longmolecules that carry genetic information; (ii) aprotein coat, called the capsid, which surrounds and protects the genetic material; and in some cases (iii) an envelope of lipidsthat surrounds the protein coat. The shapes of these virus particles range from simplehelical and icosahedral forms for some virus species to more complex structures for others. Most virus species have virions that are too small to be seen with an optical microscope. The average virion is about one one-hundredth the size of the average bacterium.
The origins of viruses in the evolutionary history of life are unclear: some may haveevolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity.[7] Viruses are considered by some to be a life form, because they carry genetic material, reproduce, and evolve throughnatural selection, but lack key characteristics (such as cell structure) that are generally considered necessary to count as life. Because they possess some but not all such qualities, viruses have been described as "organisms at the edge of life",[8] and as replicators.[9]
Viruses spread in many ways; viruses in plants are often transmitted from plant to plant by insects that feed on plant sap, such as aphids; viruses in animals can be carried by blood-sucking insects. These disease-bearing organisms are known as vectors.Influenza viruses are spread by coughing and sneezing. Norovirus and rotavirus, common causes of viral gastroenteritis, are transmitted by the faecal–oral route and are passed from person to person by contact, entering the body in food or water. HIV is one of several viruses transmitted through sexual contactand by exposure to infected blood. The variety of host cells that a virus can infect is called its "host range". This can be narrow, meaning a virus is capable of infecting few species, or broad, meaning it is capable of infecting many.[10]
Viral infections in animals provoke an immune response that usually eliminates the infecting virus. Immune responses can also be produced by vaccines, which confer anartificially acquired immunity to the specific viral infection. Some viruses, including those that cause AIDS and viral hepatitis, evade these immune responses and result in chronicinfections. Antibiotics have no effect on viruses, but several antiviral drugs have been developed.
Etymology
The word is from the Latin neuter vīrusreferring to poison and other noxious liquids, from 'the same Indo-European base as Sanskrit viṣa poison, Avestan vīša poison, ancient Greek ἰός poison', first attested in English in 1398 in John Trevisa's translation of Bartholomeus Anglicus's De Proprietatibus Rerum.[11][12] Virulent, from Latin virulentus(poisonous), dates to c. 1400.[13][14] A meaning of "agent that causes infectious disease" is first recorded in 1728,[12] before the discovery of viruses by Dmitri Ivanovsky in 1892. The English plural is viruses(sometimes also viri[15] or vira[16]), whereas the Latin word is a mass noun, which has noclassically attested plural (vīra is used in Neo-Latin[17]). The adjective viral dates to 1948.[18]The term virion (plural virions), which dates from 1959,[19] is also used to refer to a single, stable infective viral particle that is released from the cell and is fully capable of infecting other cells of the same type.[20]
History

Martinus Beijerinck in his laboratory in 1921
Louis Pasteur was unable to find a causative agent for rabies and speculated about a pathogen too small to be detected using a microscope.[21] In 1884, the Frenchmicrobiologist Charles Chamberland invented a filter (known today as the Chamberland filter or the Pasteur-Chamberland filter) with pores smaller than bacteria. Thus, he could pass a solution containing bacteria through the filter and completely remove them.[22] In 1892, the Russian biologist Dmitri Ivanovsky used this filter to study what is now known as the tobacco mosaic virus. His experiments showed that crushed leaf extracts from infected tobacco plants remain infectious after filtration. Ivanovsky suggested the infection might be caused by a toxin produced by bacteria, but did not pursue the idea.[23] At the time it was thought that all infectious agents could be retained by filters and grown on a nutrient medium – this was part of thegerm theory of disease.[2] In 1898, the Dutch microbiologist Martinus Beijerinck repeated the experiments and became convinced that the filtered solution contained a new form of infectious agent.[24] He observed that the agent multiplied only in cells that were dividing, but as his experiments did not show that it was made of particles, he called it acontagium vivum fluidum (soluble living germ) and re-introduced the word virus. Beijerinck maintained that viruses were liquid in nature, a theory later discredited by Wendell Stanley, who proved they were particulate.[23] In the same year Friedrich Loeffler and Paul Frosch passed the first animal virus – agent of foot-and-mouth disease (aphthovirus) – through a similar filter.[25]
In the early 20th century, the English bacteriologist Frederick Twort discovered a group of viruses that infect bacteria, now called bacteriophages[26] (or commonlyphages), and the French-Canadian microbiologist Félix d'Herelle described viruses that, when added to bacteria on anagar plate, would produce areas of dead bacteria. He accurately diluted a suspension of these viruses and discovered that the highest dilutions (lowest virus concentrations), rather than killing all the bacteria, formed discrete areas of dead organisms. Counting these areas and multiplying by the dilution factor allowed him to calculate the number of viruses in the original suspension.[27] Phages were heralded as a potential treatment for diseases such astyphoid and cholera, but their promise was forgotten with the development of penicillin. The study of phages provided insights into theswitching on and off of genes, and a useful mechanism for introducing foreign genes into bacteria.
By the end of the 19th century, viruses were defined in terms of their infectivity, their ability to be filtered, and their requirement for living hosts. Viruses had been grown only in plants and animals. In 1906, Ross Granville Harrisoninvented a method for growing tissue inlymph, and, in 1913, E. Steinhardt, C. Israeli, and R. A. Lambert used this method to growvaccinia virus in fragments of guinea pig corneal tissue.[28] In 1928, H. B. Maitland and M. C. Maitland grew vaccinia virus in suspensions of minced hens' kidneys. Their method was not widely adopted until the 1950s, when poliovirus was grown on a large scale for vaccine production.[29]
Another breakthrough came in 1931, when the American pathologist Ernest William Goodpasture and Alice Miles Woodruff grew influenza and several other viruses in fertilised chickens' eggs.[30] In 1949, John Franklin Enders, Thomas Weller, and Frederick Robbins grew polio virus in cultured human embryo cells, the first virus to be grown without using solid animal tissue or eggs. This work enabled Jonas Salk to make an effective polio vaccine.[31]
The first images of viruses were obtained upon the invention of electron microscopy in 1931 by the German engineers Ernst Ruskaand Max Knoll.[32] In 1935, American biochemist and virologist Wendell Meredith Stanley examined the tobacco mosaic virus and found it was mostly made of protein.[33] A short time later, this virus was separated into protein and RNA parts.[34] The tobacco mosaic virus was the first to be crystallisedand its structure could therefore be elucidated in detail. The first X-ray diffraction pictures of the crystallised virus were obtained by Bernal and Fankuchen in 1941. On the basis of her pictures, Rosalind Franklin discovered the full structure of the virus in 1955.[35] In the same year, Heinz Fraenkel-Conrat and Robley Williams showed that purified tobacco mosaic virus RNA and its protein coat can assemble by themselves to form functional viruses, suggesting that this simple mechanism was probably the means through which viruses were created within their host cells.[36]
The second half of the 20th century was the golden age of virus discovery and most of the over 2,000 recognised species of animal, plant, and bacterial viruses were discovered during these years.[37] In 1957, equine arterivirus and the cause of Bovine virus diarrhoea (a pestivirus) were discovered. In 1963, the hepatitis B virus was discovered byBaruch Blumberg,[38] and in 1965, Howard Temin described the first retrovirus. Reverse transcriptase, the enzyme that retroviruses use to make DNA copies of their RNA, was first described in 1970, independently byHoward Martin Temin and David Baltimore.[39]In 1983 Luc Montagnier's team at the Pasteur Institute in France, first isolated the retrovirus now called HIV.[40] In 1989 Michael Houghton's team at Chiron Corporationdiscovered Hepatitis C.[41][42]
Origins
Viruses are found wherever there is life and have probably existed since living cells first evolved.[43] The origin of viruses is unclear because they do not form fossils, somolecular techniques have been used to compare the DNA or RNA of viruses and are a useful means of investigating how they arose.[44] In addition, viral genetic material may occasionally integrate into the germlineof the host organisms, by which they can be passed on vertically to the offspring of the host for many generations. This provides an invaluable source of information forpaleovirologists to trace back ancient viruses that have existed up to millions of years ago. There are three main hypotheses that aim to explain the origins of viruses:[45][46]
- Regressive hypothesis
- Viruses may have once been small cells that parasitised larger cells. Over time, genes not required by their parasitism were lost. The bacteria rickettsia and chlamydiaare living cells that, like viruses, can reproduce only inside host cells. They lend support to this hypothesis, as their dependence on parasitism is likely to have caused the loss of genes that enabled them to survive outside a cell. This is also called the degeneracy hypothesis,[47][48] orreduction hypothesis.[49]
- Cellular origin hypothesis
- Some viruses may have evolved from bits of DNA or RNA that "escaped" from the genes of a larger organism. The escaped DNA could have come from plasmids(pieces of naked DNA that can movebetween cells) or transposons (molecules of DNA that replicate and move around to different positions within the genes of the cell).[50] Once called "jumping genes", transposons are examples of mobile genetic elements and could be the origin of some viruses. They were discovered in maize by Barbara McClintock in 1950.[51]This is sometimes called the vagrancy hypothesis,[47][52] or the escape hypothesis.[49]
- Co-evolution hypothesis
- This is also called the virus-first hypothesis[49] and proposes that viruses may have evolved from complex molecules of protein and nucleic acid at the same time as cells first appeared on Earth and would have been dependent on cellular life for billions of years. Viroids are molecules of RNA that are not classified as viruses because they lack a protein coat. They have characteristics that are common to several viruses and are often called subviral agents.[53] Viroids are important pathogens of plants.[54] They do not code for proteins but interact with the host cell and use the host machinery for their replication.[55] Thehepatitis delta virus of humans has an RNAgenome similar to viroids but has a protein coat derived from hepatitis B virus and cannot produce one of its own. It is, therefore, a defective virus. Although hepatitis delta virus genome may replicate independently once inside a host cell, it requires the help of hepatitis B virus to provide a protein coat so that it can be transmitted to new cells.[56] In similar manner, the sputnik virophage is dependent on mimivirus, which infects the protozoanAcanthamoeba castellanii.[57] These viruses, which are dependent on the presence of other virus species in the host cell, are called satellites and may represent evolutionary intermediates of viroids and viruses.[58][59]
In the past, there were problems with all of these hypotheses: the regressive hypothesis did not explain why even the smallest of cellular parasites do not resemble viruses in any way. The escape hypothesis did not explain the complex capsids and other structures on virus particles. The virus-first hypothesis contravened the definition of viruses in that they require host cells.[49]Viruses are now recognised as ancient and as having origins that pre-date the divergence of life into the three domains.[60] This discovery has led modern virologists to reconsider and re-evaluate these three classical hypotheses.[60]
The evidence for an ancestral world of RNAcells[61] and computer analysis of viral and host DNA sequences are giving a better understanding of the evolutionary relationships between different viruses and may help identify the ancestors of modern viruses. To date, such analyses have not proved which of these hypotheses is correct.[61] It seems unlikely that all currently known viruses have a common ancestor, and viruses have probably arisen numerous times in the past by one or more mechanisms.[62]
Prions are infectious protein molecules that do not contain DNA or RNA.[63] They can cause infections such as scrapie in sheep,bovine spongiform encephalopathy ("mad cow" disease) in cattle, and chronic wasting disease in deer; in humans, prionic diseasesinclude Kuru, Creutzfeldt–Jakob disease, andGerstmann–Sträussler–Scheinker syndrome.[64] Although prions are fundamentally different from viruses and viroids, their discovery gives credence to the theory that viruses could have evolved from self-replicating molecules.[65]
Microbiology
Life properties
Opinions differ on whether viruses are a form of life, or organic structures that interact with living organisms.[66] They have been described as "organisms at the edge of life",[8]since they resemble organisms in that they possess genes, evolve by natural selection,[67]and reproduce by creating multiple copies of themselves through self-assembly. Although they have genes, they do not have a cellular structure, which is often seen as the basic unit of life. Viruses do not have their ownmetabolism, and require a host cell to make new products. They therefore cannot naturally reproduce outside a host cell[68] – although bacterial species such as rickettsia andchlamydia are considered living organisms despite the same limitation.[69][70] Accepted forms of life use cell division to reproduce, whereas viruses spontaneously assemble within cells. They differ from autonomous growth of crystals as they inherit genetic mutations while being subject to natural selection. Virus self-assembly within host cells has implications for the study of theorigin of life, as it lends further credence to the hypothesis that life could have started asself-assembling organic molecules.[1]
Structure

Diagram of how a virus capsid can be constructed using multiple copies of just two protein molecules
Structure of tobacco mosaic virus: RNA coiled in a helix of repeating protein sub-units
Structure of icosahedraladenovirus. Electron micrograph of with a cartoon to show shape
Structure of chickenpox virus. They have a lipid envelope
Structure of an icosahedralcowpea mosaic virus
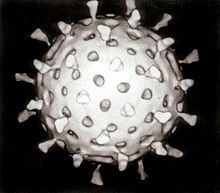
Viruses display a wide diversity of shapes and sizes, called morphologies. In general, viruses are much smaller than bacteria. Most viruses that have been studied have a diameter between 20 and 300 nanometres. Somefiloviruses have a total length of up to 1400 nm; their diameters are only about 80 nm.[71] Most viruses cannot be seen with an optical microscope so scanning and transmission electron microscopes are used to visualise them.[72] To increase the contrast between viruses and the background, electron-dense "stains" are used. These are solutions of salts of heavy metals, such astungsten, that scatter the electrons from regions covered with the stain. When virions are coated with stain (positive staining), fine detail is obscured. Negative stainingovercomes this problem by staining the background only.[73]
A complete virus particle, known as a virion, consists of nucleic acid surrounded by a protective coat of protein called a capsid. These are formed from identical protein subunits called capsomeres.[74] Viruses can have a lipid "envelope" derived from the hostcell membrane. The capsid is made from proteins encoded by the viral genome and its shape serves as the basis for morphological distinction.[75][76] Virally coded protein subunits will self-assemble to form a capsid, in general requiring the presence of the virus genome. Complex viruses code for proteins that assist in the construction of their capsid. Proteins associated with nucleic acid are known as nucleoproteins, and the association of viral capsid proteins with viral nucleic acid is called a nucleocapsid. The capsid and entire virus structure can be mechanically (physically) probed through atomic force microscopy.[77][78] In general, there are four main morphological virus types:
- Helical
- These viruses are composed of a single type of capsomere stacked around a central axis to form a helical structure, which may have a central cavity, or tube. This arrangement results in rod-shaped or filamentous virions: These can be short and highly rigid, or long and very flexible. The genetic material, in general, single-stranded RNA, but ssDNA in some cases, is bound into the protein helix by interactions between the negatively charged nucleic acid and positive charges on the protein. Overall, the length of a helical capsid is related to the length of the nucleic acid contained within it and the diameter is dependent on the size and arrangement of capsomeres. The well-studied tobacco mosaic virus is an example of a helical virus.[79]
- Icosahedral
- Most animal viruses are icosahedral or near-spherical with chiral icosahedral symmetry. A regular icosahedron is the optimum way of forming a closed shell from identical sub-units. The minimum number of identical capsomeres required for each triangular face is 3, which gives 60 for the icosahedron. Many viruses, such as rotavirus, have more than 60 capsomers and appear spherical but they retain this symmetry. To achieve this, the capsomeres at the apices are surrounded by five other capsomeres and are called pentons. Capsomeres on the triangular faces are surrounded by six others and are called hexons.[80] Hexons are in essence flat and pentons, which form the 12 vertices, are curved. The same protein may act as the subunit of both the pentamers and hexamers or they may be composed of different proteins.[81]
- Prolate
- This is an icosahedron elongated along the fivefold axis and is a common arrangement of the heads of bacteriophages. This structure is composed of a cylinder with a cap at either end.[82]
- Envelope
- Some species of virus envelop themselves in a modified form of one of the cell membranes, either the outer membrane surrounding an infected host cell or internal membranes such as nuclear membrane orendoplasmic reticulum, thus gaining an outer lipid bilayer known as a viral envelope. This membrane is studded with proteins coded for by the viral genome and host genome; the lipid membrane itself and any carbohydrates present originate entirely from the host. The influenza virus and HIV use this strategy. Most enveloped viruses are dependent on the envelope for their infectivity.[83]
- Complex
- These viruses possess a capsid that is neither purely helical nor purely icosahedral, and that may possess extra structures such as protein tails or a complex outer wall. Some bacteriophages, such asEnterobacteria phage T4, have a complex structure consisting of an icosahedral head bound to a helical tail, which may have ahexagonal base plate with protruding protein tail fibres. This tail structure acts like a molecular syringe, attaching to the bacterial host and then injecting the viral genome into the cell.[84]
The poxviruses are large, complex viruses that have an unusual morphology. The viral genome is associated with proteins within a central disc structure known as a nucleoid. The nucleoid is surrounded by a membrane and two lateral bodies of unknown function. The virus has an outer envelope with a thick layer of protein studded over its surface. The whole virion is slightly pleiomorphic, ranging from ovoid to brick shape.[85] Mimivirus is one of the largest characterised viruses, with a capsid diameter of 400 nm. Protein filaments measuring 100 nm project from the surface. The capsid appears hexagonal under an electron microscope, therefore the capsid is probably icosahedral.[86] In 2011, researchers discovered the largest then known virus in samples of water collected from the ocean floor off the coast of Las Cruces, Chile. Provisionally named Megavirus chilensis, it can be seen with a basic optical microscope.[87] In 2013, the Pandoravirusgenus was discovered in Chile and Australia, and has genomes about twice as large as Megavirus and Mimivirus.[88]
Some viruses that infect Archaea have complex structures that are unrelated to any other form of virus, with a wide variety of unusual shapes, ranging from spindle-shaped structures, to viruses that resemble hooked rods, teardrops or even bottles. Other archaeal viruses resemble the tailed bacteriophages, and can have multiple tail structures.[89]
Genome
An enormous variety of genomic structures can be seen among viral species; as a group, they contain more structural genomic diversity than plants, animals, archaea, or bacteria. There are millions of different types of viruses,[4] although only about 5,000 types have been described in detail.[3] As of September 2015, the NCBI Virus genome database has more than 75,000 complete genome sequences.[90] but there are doubtlessly many more to be discovered.[91][92]
A virus has either a DNA or an RNA genome and is called a DNA virus or an RNA virus, respectively. The vast majority of viruses have RNA genomes. Plant viruses tend to have single-stranded RNA genomes and bacteriophages tend to have double-stranded DNA genomes.[93]
Viral genomes are circular, as in thepolyomaviruses, or linear, as in theadenoviruses. The type of nucleic acid is irrelevant to the shape of the genome. Among RNA viruses and certain DNA viruses, the genome is often divided up into separate parts, in which case it is called segmented. For RNA viruses, each segment often codes for only one protein and they are usually found together in one capsid. All segments are not required to be in the same virion for the virus to be infectious, as demonstrated bybrome mosaic virus and several other plant viruses.[71]
A viral genome, irrespective of nucleic acid type, is almost always either single-strandedor double-stranded. Single-stranded genomes consist of an unpaired nucleic acid, analogous to one-half of a ladder split down the middle. Double-stranded genomes consist of two complementary paired nucleic acids, analogous to a ladder. The virus particles of some virus families, such as those belonging to the Hepadnaviridae, contain a genome that is partially double-stranded and partially single-stranded.[93]
For most viruses with RNA genomes and some with single-stranded DNA genomes, the single strands are said to be either positive-sense (called the plus-strand) or negative-sense (called the minus-strand), depending on if they are complementary to the viralmessenger RNA (mRNA). Positive-sense viral RNA is in the same sense as viral mRNA and thus at least a part of it can be immediatelytranslated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA-dependent RNA polymerase before translation. DNA nomenclature for viruses with single-sense genomic ssDNA is similar to RNA nomenclature, in that the template strandfor the viral mRNA is complementary to it (−), and the coding strand is a copy of it (+).[93]Several types of ssDNA and ssRNA viruses have genomes that are ambisense in that transcription can occur off both strands in a double-stranded replicative intermediate. Examples include geminiviruses, which are ssDNA plant viruses and arenaviruses, which are ssRNA viruses of animals.[94]
Genome size varies greatly between species. The smallest viral genomes – the ssDNA circoviruses, family Circoviridae – code for only two proteins and have a genome size of only two kilobases;[95] the largest–thepandoraviruses–have genome sizes of around two megabases which code for about 2500 proteins.[96] Virus genes rarely haveintrons and often are arranged in the genome so that they overlap.[97]
In general, RNA viruses have smaller genome sizes than DNA viruses because of a higher error-rate when replicating, and have a maximum upper size limit.[44] Beyond this limit, errors in the genome when replicating render the virus useless or uncompetitive. To compensate for this, RNA viruses often have segmented genomes – the genome is split into smaller molecules – thus reducing the chance that an error in a single-component genome will incapacitate the entire genome. In contrast, DNA viruses generally have larger genomes because of the high fidelity of their replication enzymes.[98] Single-strand DNA viruses are an exception to this rule, as mutation rates for these genomes can approach the extreme of the ssRNA virus case.[99]
Genetic mutation
How antigenic shift, or reassortment, can result in novel and highly pathogenic strains of human flu
Viruses undergo genetic change by several mechanisms. These include a process calledantigenic drift where individual bases in the DNA or RNA mutate to other bases. Most of these point mutations are "silent" – they do not change the protein that the gene encodes – but others can confer evolutionary advantages such as resistance to antiviral drugs.[100][101] Antigenic shift occurs when there is a major change in the genome of the virus. This can be a result of recombination orreassortment. When this happens with influenza viruses, pandemics might result.[102]RNA viruses often exist as quasispecies or swarms of viruses of the same species but with slightly different genome nucleoside sequences. Such quasispecies are a prime target for natural selection.[103]
Segmented genomes confer evolutionary advantages; different strains of a virus with a segmented genome can shuffle and combine genes and produce progeny viruses or (offspring) that have unique characteristics. This is called reassortment or viral sex.[104]
Genetic recombination is the process by which a strand of DNA is broken and then joined to the end of a different DNA molecule. This can occur when viruses infect cells simultaneously and studies of viral evolutionhave shown that recombination has been rampant in the species studied.[105]Recombination is common to both RNA and DNA viruses.[106][107]
Replication cycle
Viral populations do not grow through cell division, because they are acellular. Instead, they use the machinery and metabolism of a host cell to produce multiple copies of themselves, and they assemble in the cell.
Some bacteriophages inject their genomesinto bacterial cells (not to scale)
The life cycle of viruses differs greatly between species but there are six basicstages in the life cycle of viruses:[108]
Attachment is a specific binding between viral capsid proteins and specific receptors on the host cellular surface. This specificity determines the host range of a virus. For example, HIV infects a limited range of humanleucocytes. This is because its surface protein, gp120, specifically interacts with theCD4 molecule – a chemokine receptor – which is most commonly found on the surface of CD4+ T-Cells. This mechanism has evolved to favour those viruses that infect only cells in which they are capable of replication. Attachment to the receptor can induce the viral envelope protein to undergo changes that results in the fusion of viral and cellular membranes, or changes of non-enveloped virus surface proteins that allow the virus to enter.
Penetration follows attachment: Virions enter the host cell through receptor-mediatedendocytosis or membrane fusion. This is often called viral entry. The infection of plant and fungal cells is different from that of animal cells. Plants have a rigid cell wall made of cellulose, and fungi one of chitin, so most viruses can get inside these cells only after trauma to the cell wall.[109] Nearly all plant viruses (such as tobacco mosaic virus) can also move directly from cell to cell, in the form of single-stranded nucleoprotein complexes, through pores calledplasmodesmata.[110] Bacteria, like plants, have strong cell walls that a virus must breach to infect the cell. Given that bacterial cell walls are much thinner than plant cell walls due to their much smaller size, some viruses have evolved mechanisms that inject their genome into the bacterial cell across the cell wall, while the viral capsid remains outside.[111]
Uncoating is a process in which the viral capsid is removed: This may be by degradation by viral enzymes or host enzymes or by simple dissociation; the end-result is the releasing of the viral genomic nucleic acid.
Replication of viruses involves primarily multiplication of the genome. Replication involves synthesis of viral messenger RNA (mRNA) from "early" genes (with exceptions for positive sense RNA viruses), viral protein synthesis, possible assembly of viral proteins, then viral genome replication mediated by early or regulatory protein expression. This may be followed, for complex viruses with larger genomes, by one or more further rounds of mRNA synthesis: "late" gene expression is, in general, of structural or virion proteins.
Assembly – Following the structure-mediated self-assembly of the virus particles, some modification of the proteins often occurs. In viruses such as HIV, this modification (sometimes called maturation) occurs afterthe virus has been released from the host cell.[112]
Release – Viruses can be released from the host cell by lysis, a process that kills the cell by bursting its membrane and cell wall if present: This is a feature of many bacterial and some animal viruses. Some viruses undergo a lysogenic cycle where the viral genome is incorporated by genetic recombination into a specific place in the host's chromosome. The viral genome is then known as a "provirus" or, in the case of bacteriophages a "prophage".[113] Whenever the host divides, the viral genome is also replicated. The viral genome is mostly silent within the host. At some point, the provirus or prophage may give rise to active virus, which may lyse the host cells.[114] Enveloped viruses (e.g., HIV) typically are released from the host cell by budding. During this process the virus acquires its envelope, which is a modified piece of the host's plasma or other, internal membrane.[115]
Genome replication
The genetic material within virus particles, and the method by which the material is replicated, varies considerably between different types of viruses.
- DNA viruses
- The genome replication of most DNA viruses takes place in the cell's nucleus. If the cell has the appropriate receptor on its surface, these viruses enter the cell sometimes by direct fusion with the cell membrane (e.g., herpesviruses) or – more usually – by receptor-mediated endocytosis. Most DNA viruses are entirely dependent on the host cell's DNA and RNA synthesising machinery, and RNA processing machinery. Viruses with larger genomes may encode much of this machinery themselves. In eukaryotes the viral genome must cross the cell's nuclear membrane to access this machinery, while in bacteria it need only enter the cell.[116]
- RNA viruses
- Replication usually takes place in thecytoplasm. RNA viruses can be placed into four different groups depending on their modes of replication. The polarity (whether or not it can be used directly by ribosomes to make proteins) of single-stranded RNA viruses largely determines the replicative mechanism; the other major criterion is whether the genetic material is single-stranded or double-stranded. All RNA viruses use their own RNA replicaseenzymes to create copies of their genomes.[117]
- Reverse transcribing viruses
- These have ssRNA (Retroviridae,Metaviridae, Pseudoviridae) or dsDNA (Caulimoviridae, and Hepadnaviridae) in their particles. Reverse transcribing viruses with RNA genomes (retroviruses), use a DNA intermediate to replicate, whereas those with DNA genomes (pararetroviruses) use an RNA intermediate during genome replication. Both types use a reverse transcriptase, or RNA-dependent DNA polymerase enzyme, to carry out the nucleic acid conversion. Retroviruses integrate the DNA produced by reverse transcription into the host genome as a provirus as a part of the replication process; pararetroviruses do not, although integrated genome copies of especially plant pararetroviruses can give rise to infectious virus.[118] They are susceptible to antiviral drugs that inhibit the reverse transcriptase enzyme, e.g.zidovudine and lamivudine. An example of the first type is HIV, which is a retrovirus. Examples of the second type are theHepadnaviridae, which includes Hepatitis B virus.[119]
Effects on the host cell
The range of structural and biochemical effects that viruses have on the host cell is extensive.[120] These are called cytopathic effects.[121] Most virus infections eventually result in the death of the host cell. The causes of death include cell lysis, alterations to the cell's surface membrane and apoptosis.[122]Often cell death is caused by cessation of its normal activities because of suppression by virus-specific proteins, not all of which are components of the virus particle.[123]
Some viruses cause no apparent changes to the infected cell. Cells in which the virus islatent and inactive show few signs of infection and often function normally.[124] This causes persistent infections and the virus is often dormant for many months or years. This is often the case with herpes viruses.[125][126]Some viruses, such as Epstein–Barr virus, can cause cells to proliferate without causing malignancy,[127] while others, such aspapillomaviruses, are established causes of cancer.[128]
Host range
Viruses are by far the most abundant biological entities on Earth and they outnumber all the others put together.[129]They infect all types of cellular life including animals, plants, bacteria and fungi.[3] Different types of viruses can infect only a limited range of hosts and many are species-specific. Some, such as smallpox virus for example, can infect only one species – in this case humans,[130] and are said to have a narrowhost range. Other viruses, such as rabies virus, can infect different species of mammals and are said to have a broad range.[131] The viruses that infect plants are harmless to animals, and most viruses that infect other animals are harmless to humans.[132] The host range of some bacteriophages is limited to a single strain of bacteria and they can be used to trace the source of outbreaks of infections by a method called phage typing.[133]
Comments
Post a Comment